Overview of Enterobacterales: Key Pathogens, Identification, and Clinical Significance
Overview of Enterobacterales: The Medically Important Family of Gram-Negative Bacteria
In clinical microbiology, few bacterial groups are as ubiquitous, diverse, and clinically significant as the Enterobacterales.
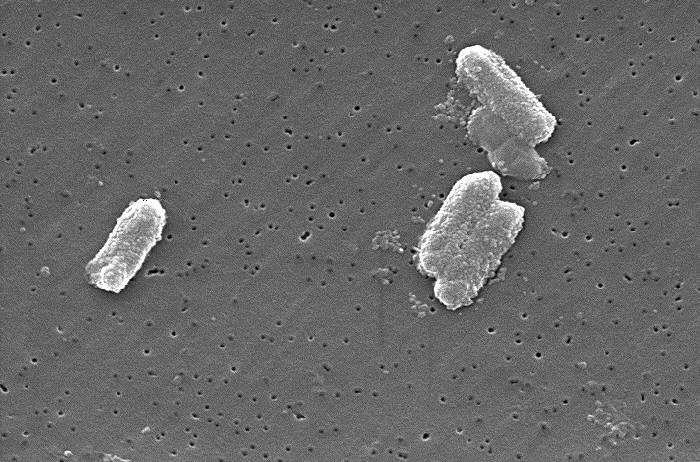
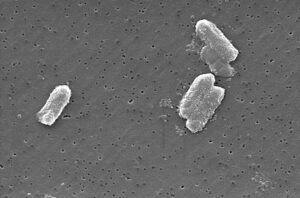
This order of Gram-negative, rod-shaped, facultatively anaerobic bacteria includes some of the most common causes of human infection—from urinary tract infections (UTIs) and gastroenteritis to life-threatening sepsis and pneumonia. But it also includes harmless gut commensals and environmental species.
Understanding Enterobacterales is essential for accurate diagnosis, effective treatment, and antimicrobial stewardship. Let’s break down everything you need to know.
What Are Enterobacterales?
Formerly classified under the family Enterobacteriaceae, the group was reorganized in 2016 based on genomic data. Today, Enterobacterales is an order containing 7 families, with the Enterobacteriaceae family still housing the majority of clinically relevant genera.
Key characteristics:
- Gram-negative rods (bacilli)
- Facultative anaerobes (can grow with or without oxygen)
- Ferment glucose (most produce acid ± gas)
- Oxidase-negative (a key test to distinguish from Pseudomonas)
- Motile or non-motile (many have peritrichous flagella)
- Reduce nitrates to nitrites
These traits make them well-suited to thrive in the human gut, soil, water, and hospital environments.
Clinically Important Genera in Enterobacterales
While over 30 genera exist, these are the most common pathogens in human medicine:
| GENUS | COMMON INFECTIONS | NOTABLE FEATURES |
|---|---|---|
| Escherichia | UTIs, sepsis, neonatal meningitis, traveler’s diarrhea | E. coliis the #1 cause of community-acquired UTIs |
| Klebsiella | Pneumonia (especially in alcoholics), UTIs, wound infections | Mucoid colonies; associated withESBLandcarbapenemaseproduction |
| Enterobacter | Hospital-acquired pneumonia, catheter-related infections | Intrinsically resistant to ampicillin; can developAmpC β-lactamase |
| Citrobacter | UTIs, sepsis, neonatal meningitis | Can mimicSalmonellabiochemically |
| Serratia | Respiratory infections, UTIs, endocarditis | Produces red pigment (prodigiosin) at room temp; often multidrug-resistant |
| Proteus | UTIs (especially with stones), wound infections | Swarming motilityon agar; urease-positive → alkalinizes urine |
| Morganella | UTIs, wound infections | Urease-positive; often resistant to tetracyclines |
| Providencia | UTIs (in catheterized patients), traveler’s diarrhea | Associated with summer diarrhea outbreaks |
| Salmonella | Gastroenteritis, typhoid fever, bacteremia | Non-lactose fermenter; motile; >2,600 serotypes |
| Shigella | Bacillary dysentery (bloody diarrhea) | Non-motile; invades colonic mucosa; low infectious dose |
💡 Note: Yersinia (including Y. pestis, cause of plague) is also part of this order but less common in routine clinical labs.
Why Are Enterobacterales So Clinically Relevant?
- Ubiquity: They colonize the human gut (e.g., E. coli) and are easily transmitted via fecal-oral route or contaminated surfaces.
- Opportunism: Many cause disease when host defenses are compromised (e.g., ICU patients, diabetics, catheterized individuals).
- Antimicrobial Resistance (AMR): Enterobacterales are leading drivers of multidrug resistance, including:
- ESBLs (Extended-Spectrum β-Lactamases)
- Carbapenemases (e.g., KPC, NDM, OXA-48)
- AmpC β-lactamases (often inducible in Enterobacter, Citrobacter, Serratia)
- Healthcare Burden: They account for ~50% of all Gram-negative hospital infections worldwide.
Laboratory Identification: From Gram Stain to MALDI-TOF
Step 1: Gram Stain
- Reveals Gram-negative rods—immediate clue.
Step 2: Culture on Selective Media
- MacConkey agar: Differentiates lactose fermenters (pink colonies: E. coli, Klebsiella) vs. non-fermenters (colorless: Salmonella, Shigella, Proteus).
- XLD or Hektoen agar: Used for stool cultures to isolate Salmonella/Shigella.
Step 3: Biochemical Testing
Traditional tests include:
- Indole (E. coli = positive)
- Urease (Proteus = positive)
- H₂S production (Salmonella = positive on TSI)
- Motility (Shigella = non-motile)
Step 4: Modern Methods
- MALDI-TOF MS: Rapid, accurate genus/species ID from colonies
- Molecular PCR panels: Detect pathogens + resistance genes directly from specimens (e.g., stool, blood)
Antimicrobial Resistance: A Growing Crisis
Enterobacterales are at the epicenter of the AMR pandemic:
- ESBL-producing strains: Resistant to penicillins, cephalosporins, and aztreonam. Treated with carbapenems (though resistance is rising).
- Carbapenem-resistant Enterobacterales (CRE): Often pan-resistant. Treatment options include:
- Ceftazidime-avibactam
- Meropenem-vaborbactam
- Plazomicin (aminoglycoside)
- Cefiderocol (siderophore cephalosporin)
- Colistin resistance: Mediated by mcr genes—now a global concern.
🚨 Infection Control Alert: CRE are “urgent threats” (CDC). Strict contact precautions and screening are essential in hospitals.
Prevention and Public Health
- Hand hygiene: Most effective way to prevent transmission
- Safe food/water: Critical for Salmonella, Shigella, pathogenic E. coli (e.g., O157:H7)
- Antibiotic stewardship: Avoid unnecessary cephalosporins to reduce ESBL selection
- Surveillance: Track resistance patterns locally and globally (e.g., via WHONET)
Final Thought: Respect the Rod
Enterobacterales are masters of adaptation. They live peacefully in our guts, cause mild diarrhea, or trigger deadly sepsis—often depending on host factors and bacterial virulence.
For microbiologists and clinicians, staying ahead means:
- Knowing the key genera and their resistance patterns
- Using rapid diagnostics wisely
- Championing stewardship and infection control
Because in the world of Gram-negative rods, complacency can be fatal.
Want More Clinical Microbiology Insights?
At ClinicalMicrobiology.org, we deliver expert, evidence-based coverage of pathogens, diagnostics, and antimicrobial resistance—designed for real-world practice.
👉 Subscribe today for weekly updates on bacterial ID, emerging threats, and lab best practices.
Note: This article reflects current taxonomy (ICSP 2016+) and clinical guidelines (CLSI, CDC, IDSA). All content is original and intended for educational use.




Post Comment